Gene expression is a precisely regulated process that dictates when, where, and under what conditions specific genes are activated. This control underpins normal development, cellular function, and the ability to adapt to environmental changes. At the transcriptional level, regulation is driven by elements such as transcription factors, enhancers, and silencers, which respond dynamically to internal signals like cell cycle progression or hormonal cues, as well as external factors such as temperature shifts or stress. The resulting transcripts—including mRNA and diverse non-coding RNAs—play pivotal roles in shaping cellular behavior and determining functional outcomes.
RNA analysis, in particular, provides a direct window into gene activity, signaling pathways, and disease mechanisms. As a result, gene expression profiling has become a cornerstone of modern molecular biology and biotechnology, enabling precise monitoring of biological responses across cell types, tissues, and organisms.
Vazyme delivers a fully integrated, high-performance solution for gene expression analysis—covering every step from RNA extraction to real-time quantitative PCR (RT-qPCR). Our portfolio includes:
RNA extraction kits (manual and automated), optimized for a wide range of sample types including tissue, blood, plant, and degraded clinical materials;
Robust reverse transcription systems, specially engineered for superior performance with low-input or partially degraded RNA samples. Powered by our HiScript IV Reverse Transcriptase, these systems feature enhanced sensitivity, high processivity, and excellent cDNA yield—ensuring accurate and full-length transcription even at sub-nanogram RNA levels
Ultra-sensitive qPCR reagents that ensure clean amplification curves, excellent reproducibility, and consistent performance across targets of varying abundance.
By combining simplified workflows, proven robustness, and broad compatibility, Vazyme empowers researchers to generate reproducible, publication-ready data—faster and with greater confidence.
Background
Gene expression profiling is an essential approach in modern life science research, supporting applications from basic biology to disease diagnostics, drug discovery, and cell therapy development. As biological systems become more complex and sample types increasingly diverse, researchers face mounting challenges in generating high-quality and reproducible data.
Key challenges include:
Low RNA yields, especially in rare cell populations, biopsies, or single-cell studies
RNA degradation, commonly encountered in clinical samples or long-term stored specimens
Inhibitory substances from complex matrices like plant tissues, blood, or FFPE samples
Reverse transcription bias, which may compromise accuracy in quantifying transcript abundance
Workflow inefficiencies, including time-consuming RNA purification and inconsistent RT-qPCR setups
These obstacles often lead to reduced sensitivity, poor reproducibility, or even false-negative results—undermining data integrity and increasing experimental cost.
To address these pain points, gene expression analysis demands robust solutions that combine:
Efficient and clean RNA extraction, adaptable to various sample types and throughput needs
High-fidelity reverse transcription, even from degraded or low-input RNA
Sensitive, specific qPCR detection, capable of resolving subtle gene expression differences
Seamless integration across steps, to minimize technical variability and hands-on time
Vazyme’s gene expression workflow is designed to address these evolving demands, giving researchers the reliability, sensitivity, and flexibility necessary for high-quality transcriptional analysis across a wide spectrum of samples and research applications.
Reliable gene expression analysis begins with high-quality, high-integrity RNA. Vazyme’s RNA extraction solutions deliver consistent purity across a wide range of samples—from delicate plasma to polysaccharide-rich plant tissues—whether using manual or automated workflows. With dependable performance from the start, you can focus on advancing your research with confidence.
To meet the varied demands of gene expression studies, we offer optimized nucleic acid extraction kits for both animal- and plant-derived samples, available in manual and automated formats:
1. Animal-derived samples (e.g., cells and tissues)
Manual extraction: FastPure Cell/Tissue Total RNA Isolation Kit V2 (Vazyme #RC112)– trusted by researchers for its high yield and purity, particularly suited for small-scale applications or cases requiring high RNA integrity.
Automated extraction: VAMNE Magnetic Cell/Tissue Total RNA Kit (Vazyme #RMA101-C2) – ideal for high-throughput needs with consistent performance across different tissue types.
Vazyme #RC112 ('V' in the following figures) and commercially available related products (from suppliers A, B, C, D, E ,F and G respectively) were used to extract total RNA from the rat liver tissue (10 mg) and HEK 293 cell (1×106) samples according to the respective Instructions for Use. Figure A is the agarose gel electrophoretogram of the extracted RNA, while Figures B and C are the statistical charts of RNA concentration and purity results. As shown in the figures, RNA extracted by Vazyme #RC112 has good integrity, high yield and high purity.
Vazyme RMA101-C2 and commercially available comparable products (Supplier TH and Supplier Ti) were used to extract total RNA from mouse heart, rat brain, rat lung, and rat liver samples according to the respective manuals (Figure A-D). RNA extracted and purified by RMA101-C2 was analyzed by agarose gel electrophoresis (Figure E).
Vazyme’s RNA extraction portfolio is designed to deliver consistent purity and integrity across both animal and plant samples, ensuring reliable results from the very first step of gene expression analysis. For animal-derived materials, we offer manual and automated kits that combine high yield, purity, and reproducibility. For plant-derived materials, our solutions handle a broad spectrum of tissues—including those rich in polysaccharides and polyphenols—maintaining high compatibility with downstream RT-qPCR and supporting both small-scale and high-throughput workflows.
2. Plant samples (e.g., leaves, roots, seeds)
Manual extraction: FastPure Universal Plant Total RNA Isolation Kit (Vazyme #RC411) – designed for efficient RNA extraction from a wide variety of plant tissues, including those rich in polysaccharides and polyphenols, ensuring high compatibility with downstream RT-qPCR.
Automated extraction: VAMNE Magnetic Universal Plant Total RNA Kit (Vazyme #RMA4101) –optimized for robust performance on complex plant matrices within automated workflows, delivering consistent yields and purity.
RNA was extracted from various plant types—including common plants, fruit pulp, and tissues rich in polysaccharides and polyphenols—as well as from fungal samples using Vazyme’s FastPure Universal Plant Total RNA Isolation Kit (Vazyme #RC411). Agarose gel electrophoresis confirmed that RC411 delivers high-quality RNA and is compatible with a wide range of plant materials and selected fungal samples.
Featured Products

Animal-derived Samples

Plant Samples
Reliable cDNA synthesis is critical for accurate gene expression analysis, particularly when working with low-input, partially degraded, or structurally complex RNA. Vazyme’s reverse transcription solutions are powered by HiScript IV Reverse Transcriptase, our 4th generation enzyme engineered from M-MLV, representing a significant advancement in RT technology.
Key performance features:
Extended synthesis capability – Produces full-length cDNA up to 20 kb, ensuring complete coverage of long transcripts.
Rapid reaction kinetics – Completes cDNA synthesis in as little as 5 minutes, reducing turnaround time without compromising quality.
High reverse transcription efficiency – Optimized for challenging inputs, including FFPE samples and clinical RNA.
Exceptional thermal stability – Operates effectively at elevated temperatures (37–50 °C) to resolve complex secondary structures and high-GC templates.
No RNase H activity – Maintains the integrity of synthesized cDNA for downstream applications.
For qPCR workflows, HiScript IV All-in-One Ultra RT SuperMix for qPCR (Vazyme #R433) integrates enzyme, buffer, and optimized components into a single-tube format, improving reproducibility, reducing pipetting steps, and delivering faster, high-efficiency cDNA synthesis.
Superior Efficiency, Broad Compatibility
R433 consistently outperforms across diverse sample types with lower CT values and robust cDNA yields
Reverse transcription reactions were conducted using 100 ng of RNA from animal cells, plant tissues, and animal tissues as templates with Vazyme #R433, and other commercially available reverse transcription reagents (Th、Tr、TA、TI), following the recommended protocols for each product. The resulting cDNA underwent quantitative analysis of multiple genes through qPCR. The results indicate that Vazyme #R433 consistently yielded lower CT values across different testing systems, indicating superior reverse transcription efficincy and excellent compatibility.
Streamlined Workflow, No Compromise on Performance
R433 delivers reliable reverse transcription in just 5 minutes or with extended time.
Featured Products

All-in-One RT SuperMix

RT SuperMix for qPCR

Universal RT Kit
Accurate and reproducible gene expression quantification relies on precise amplification control. Vazyme’s real-time PCR reagents incorporate dual-antibody modified hot-start technology, delivering high specificity, stability, and suitability for automated workflows.
Powered by our proprietary BioSmart enzyme screening platform, which combines directed evolution with computer-aided design, we have developed a hot-start DNA polymerase modified with two monoclonal antibodies. This dual mechanism simultaneously inhibits polymerase catalytic activity and template binding at room temperature. Compared to conventional hot-start approaches, the dual-antibody strategy ensures more complete enzyme inactivation, significantly reducing nonspecific amplification during pre-amplification and improving assay accuracy.
Dual-blocking mechanism – Significantly reduces nonspecific activity by inhibiting both polymerase function and template interaction before thermal activation.
Ultra-low background noise – Produces clean amplification curves and single melting peaks for confident data interpretation.
Rapid activation – Fully activates during the initial high-temperature denaturation step, with no prolonged preheating required.
Excellent stability – Maintains reaction integrity for over 72 hours at room temperature, ideal for automated and pre-mixed workflows.
Broad compatibility – Performs reliably across diverse sample types, supporting both research and clinical needs.
Building on this enzyme innovation, SupRealQ Ultra Hunter SYBR qPCR Master Mix (U+) (Vazyme #Q713) has been developed to meet the demands of fast cycling, challenging templates, and low-abundance targets—providing exceptional sensitivity and specificity for the most demanding gene expression studies.
Sensitive to the Signal. Immune to the Noise
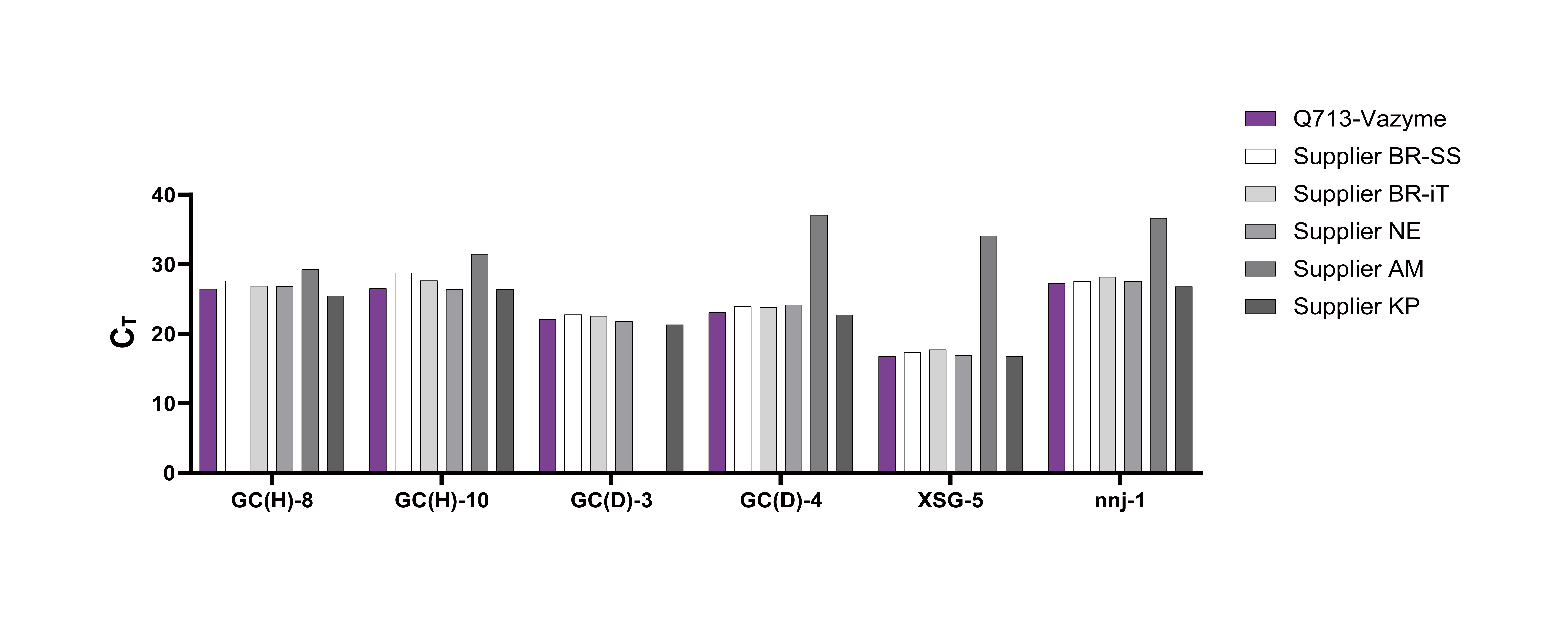
Q713 delivers exceptional sensitivity across diverse templates, consistently matching or surpassing global benchmarks. It maintains reliable performance across a range of GC contents and expression levels, making it ideal for challenging templates and low-copy targets. Powered by dual-antibody hot-start enzyme technology, Q713 ensures clean, specific amplification with minimal nonspecific signals, giving researchers confidence in accurate and reproducible gene expression measurements.
Comparative testing of Q713 PCR reagent against five international competitors across multiple cDNA samples demonstrated that Q713 consistently achieved equal or superior sensitivity, as indicated by lower or comparable Ct values. These results confirm that Q713 matches or exceeds the performance of leading global products.
Unmatched Compatibility, Uncompromised Results
Q713 is designed for robust performance across challenging samples, including low-abundance transcripts, degraded RNA, and templates with extreme GC content. Comparative tests with leading global reagents show Q713 consistently delivers reliable amplification, achieving lower or comparable Ct values across high-GC, low-GC, and long-fragment targets.
Featured Products
Next-Gen qPCR

Star qPCR
Whether you're processing a few samples or running high-throughput assays, our extraction and qPCR platforms are designed to streamline your workflow and deliver consistent results.
RNA Extraction Platform
VNP-32 Nucleic Acid Extractor – flexible, efficient, and automation-ready
Flexible throughput: Supports 1, 8, 16, or 32 samples in a single run to meet both low- and high-volume demands
High-efficiency extraction: Optimized for use with Vazyme magnetic RNA kits for reliable yield and purity
Compact and user-friendly: Intuitive touchscreen interface with pre-set and customizable protocols
Cross-contamination prevention: Enclosed design with magnetic bead separation ensures sample integrity
VNP-32 is ideal for laboratories seeking scalability, consistency, and automation in RNA extraction workflows, from research applications to routine diagnostics.
Real-time PCR System
FMR-5S - Smart,Speed, Stability, Sensitivity, Screen.
FMR-5S Real-Time PCR System delivers exceptional performance in gene expression analysis, combining intelligent design with high speed and sensitivity. Engineered for both research flexibility and routine reliability, it seamlessly supports the full qPCR workflow.
12.1" HD Touchscreen: Large, intuitive interface for streamlined experiment setup and real-time visualization.
Smart Gradient Optimization:12-column independent temperature control enables rapid condition screening in a single run.
Ultra-Fast Workflow: Fast ramping (up to 8°C/s) and full-plate fluorescence detection in just 5 seconds boost efficiency.
High Sensitivity & Low Background: Optimized optics deliver clear, strong signals—even for low-abundance targets.
Long-Term Stability: LED-based optical system with no maintenance required, ensuring reliable performance over time.
Automation-Ready Design: Supports auto-sample tray, multi-instrument linkage, and seamless integration into high-throughput workflows.
Versatile Analysis Modes: Covers absolute/relative quantification, genotyping, HRM, MMCA, protein melt, and more.

Automatic Nucleic Acids Extraction Instrument

FMR-5S Smart Quant System
Download our brochures for practical tools and insights to enhance your workflow efficiently and precisely.
Wang, G., Peng, S., Reyes Mendez, M., Keramidas, A., Castellano, D., Wu, K., Han, W., Tian, Q., Dong, L., Li, Y., & Lu, W. (2024). The TMEM132B-GABAA receptor complex controls alcohol actions in the brain. Cell, 187(23), 6649–6668.e35. https://doi.org/10.1016/j.cell.2024.09.006
Yin, W., Rajvanshi, P.K., Rogers, H.M. et al. Erythropoietin regulates energy metabolism through EPO-EpoR-RUNX1 axis. Nat Commun 15, 8114 (2024). https://doi.org/10.1038/s41467-024-52352-z
Gritti, I., Wan, J., Weeresekara, V., Vaz, J. M., Tarantino, G., Bryde, T. H., Vijay, V., Kammula, A. V., Kattel, P., Zhu, S., Vu, P., Chan, M., Wu, M. J., Gordan, J. D., Patra, K. C., Silveira, V. S., Manguso, R. T., Wein, M. N., Ott, C. J., Qi, J., … Bardeesy, N. (2025). DNAJB1-PRKACA Fusion Drives Fibrolamellar Liver Cancer through Impaired SIK Signaling and CRTC2/p300-Mediated Transcriptional Reprogramming. Cancer discovery, 15(2), 382–400. https://doi.org/10.1158/2159-8290.CD-24-0634
Li, L., Adamson, C., Ng, E.W.L. et al. Candida albicans Ssy1 is the extracellular sensor of gut microbiota-derived peptidoglycan fragments mediating invasive hyphal growth in the host. Nat Commun 16, 6737 (2025). https://doi.org/10.1038/s41467-025-62097-y
Zhang, Y., Shan, K.Z., Liang, P. et al. PIEZO1 drives trophoblast fusion and placental development. Nat Commun 16, 6895 (2025). https://doi.org/10.1038/s41467-025-62254-3
Liang, H., Li, F., Fang, H. et al. A novel peptide 66CTG stabilizes Myc proto-oncogene protein to promote triple-negative breast cancer growth. Sig Transduct Target Ther 10, 217 (2025). https://doi.org/10.1038/s41392-025-02298-5
A: Yes. We recommend using a highly sensitive and efficient reverse transcription system (e.g., Vazyme #R433), which is ideal for low-input RNA (<10 ng), including FFPE or clinical samples.
A: We suggest using a spectrophotometer to check A260/280 (ideal: 1.8–2.0) and A260/230 (>2.0) ratios. RNA integrity can also be evaluated by agarose gel electrophoresis or bioanalyzer (RIN > 7 is preferred).
A: Yes. These components can interfere with RNA purity. We recommend plant-specific kits (e.g., Vazyme #RC411 / #RMA4101) that efficiently remove polysaccharides and polyphenols to ensure RT-qPCR compatibility.
A: Not always. Vazyme RNA extraction kits are equipped with high-efficiency gDNA filter columns that typically eliminate genomic DNA without the need for DNase I. Additionally, the HiScript IV All-in-One Ultra RT SuperMix for qPCR (Cat# R433) includes a built-in gDNA wiper module that removes genomic DNA with up to 99.99% efficiency, ensuring reliable results even in high-sensitivity assays.
A: This could be due to non-specific amplification or primer-dimers. We suggest: ·Using a hot-start enzyme (e.g., Q713 with dual-antibody modification) ·Redesigning primers ·Optimizing annealing temperature ·Verifying template purity
A: Yes. With high-performance RT enzymes (e.g., HiScript IV, Vazyme #R433), cDNA synthesis can be completed in as little as 5 minutes—ideal for high-throughput or time-sensitive workflows.
A: Use a dual-antibody hot-start enzyme (e.g., Q713) to minimize non-specific activity during reaction setup. Also ensure reaction cleanliness and avoid template contamination via aerosols.